Genome sequencing analyses of “alien” Nazca mummies do not support an extraterrestrial origin
Context
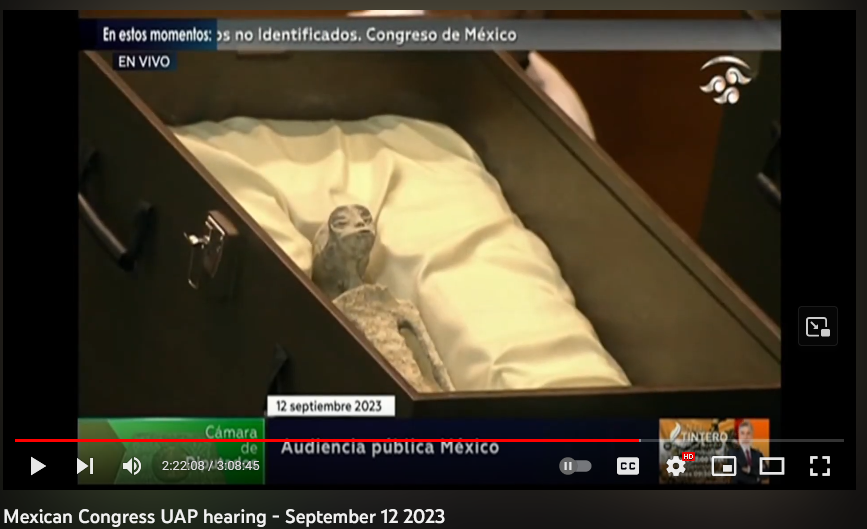
On September 12, researchers presented 2 Nazca mummies to the General Congress of the United Mexican States. They report that these mummies are definitively non-human (but humanoid) biological entities and showcased evidence from genomic studies and anatomical analyses. If their conclusions are correct, this indicates an important change in our understanding of biology and taxonomy – to say nothing of the more sensationalist claims that the mummies are extraterrestrial in origin.
José de la Cruz Ríos, Jamie Maussan, and José de Jesus Zalce Benitez presented their findings in Mexico City. Genomic reads from 3 samples have been submitted to the NIH Sequence Read Archive (SRA) by a researcher affiliated with the Universidad Nacional Autónoma de México who performed some genetic analysis presented in the hearing. The researchers say they sent the samples to multiple groups for genetic analysis, none of whom appear to have been directly involved in the hearing before the Congress.
Previously known information on the mummies
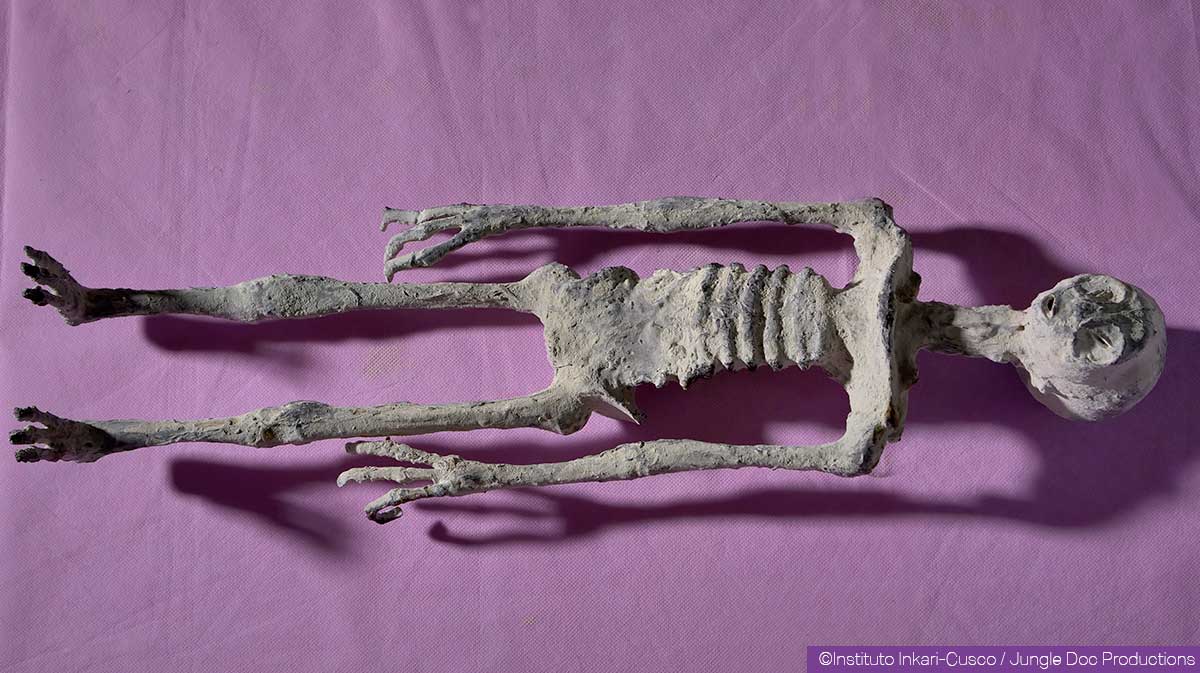
All 3 of the presenters have previously presented evidence on non-human humanoid Nazca mummies before the Congress of the Republic of Peru in 2018. Based on the testimony provided before the Mexican Congress, these seem to be a subset of the same mummies previously shown. The mummies have been extensively examined by a team who documents their results and sells DVDs with further information on www.the-alien-project.com They have 3-fingered hands and feet, elongated skulls, and spinal connections in the center of the skull.
An overview of a thorough debunking of these specimens can be accessed at https://antropogenez.ru/review/1119/. In non-exhaustive summary: X-ray experts independent of the mummy discovery group say that the specimens consist of human bones intentionally placed into an arrangement which makes no biological sense. The elongated skulls on the ‘humanoid reptile’ type, presented to the Mexican Congress, are hypothesized to be partial skulls from other mummified mammals contemporaneous to true ancient Nazca mummies.
During the presentation before General Congress of the United Mexican States, the researchers indicated that these specimens were discovered in 2015, at the same time as specimens which have already been presented in Peru. The Mexican Congress specimens are identifiable as the “humanoid reptile” type presented on the alien project website.
Here is “Alberto”, a specimen previously described as a humanoid reptile.
Here is one of the non-human humanoids presented before the Mexican Congress on September 12. The resemblance to “Alberto” is clear.
Preponderance of unaligned reads consistent with low yield extractions from ancient samples
The SRA samples provided have the same base count, GC content, and sample identifiers as samples discussed in an Abraxas Biosystems consulting report from 2018, uploaded by the Alien Project on their website.These data indicate that the Abraxas samples and SRA samples are the same – particularly the identical base count. The Abraxas Biosystems report describes sample Ancient002 (“sample 2”) and sample Ancient004 (“sample 4”) as being from different locations (bone and tissue) on the same mummy, called “Victoria”. “Victoria” is a headless humanoid mummy, and not one of the ones presented to the General Congress of the United Mexican States. Sample Ancient003 (“sample 3”) is described as a separate hand.
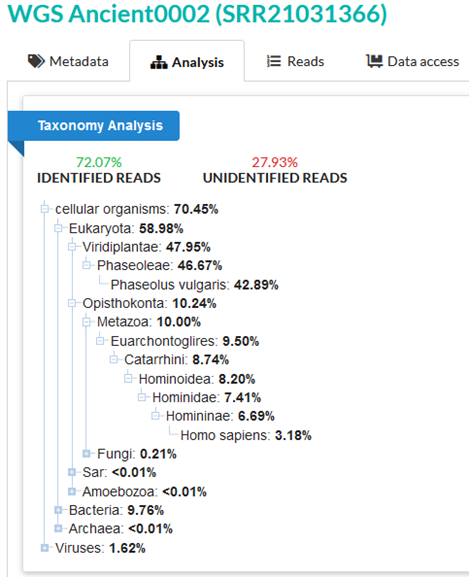
Each sample in the SRA has a BioSample accession, and all 3 samples were identified by the submitter as human. Samples Ancient002 (“sample 2”) and Ancient003 (“sample 3”) are identified as bone, and sample Ancient004 (“sample 4”) is identified as muscle tissue. GC content of the samples ranges between 39.7-46.4%, which is not inconsistent with the range of GC content in human DNA. Native SRA taxonomy analysis is available for each of the 3 samples. Sample 2’s 39.7% GC content is relatively low for human DNA, but is more typical of legumes, for example – more on this below.
42.89% of reads in sample 2 are confidently assigned to Phaseolus vulgaris, the common bean. This is most easily explained by sample contamination or construction of the putative bone fragment from a bean derivative.
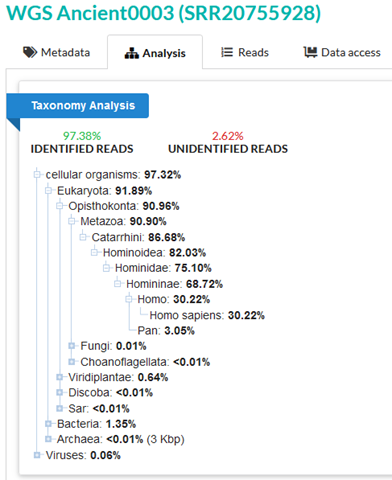
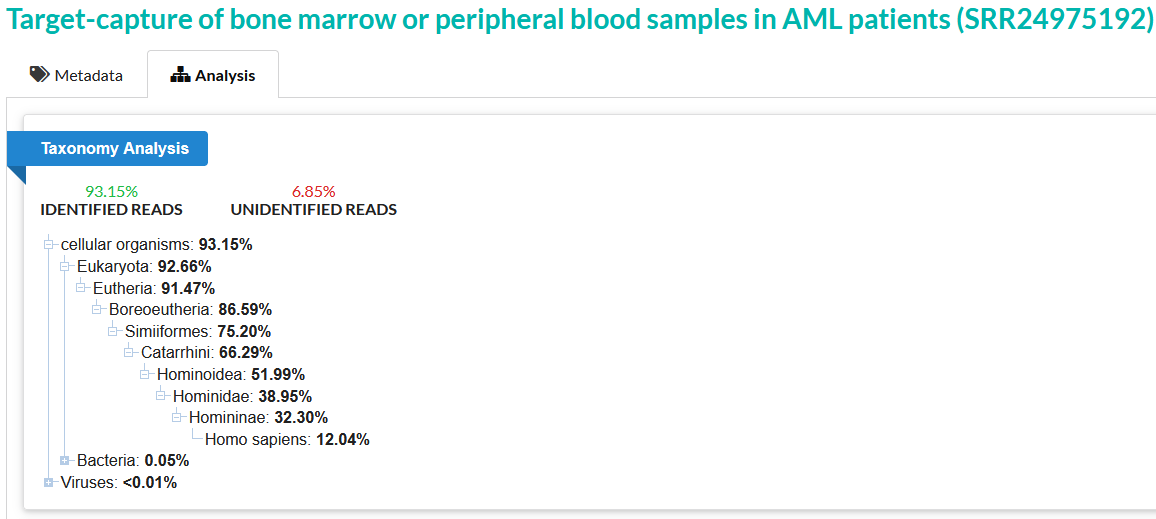
SRA taxonomy analysis confidently assigns 97.38% of the reads in sample 3 to known taxonomic categories. Only 30.22% of reads can be confidently assigned to Homo sapiens, which can initially seem like an indication of some DNA of non-human origin. However, let’s compare this to an SRA taxonomy analysis of a known high-quality human sample.
Here, we see that only 93.15% of reads can be confidently identified – this is actually lower than the percentage of identified reads in sample 3. And only 12.04% of reads are confidently assigned to Homo sapiens – much lower than the 30.22% which can be assigned in sample 3. In this context, sample 3 is almost definitively human DNA. The Abraxas report, discussed earlier, also identifies sample 3 as containing human DNA, and further specifically as a human male.
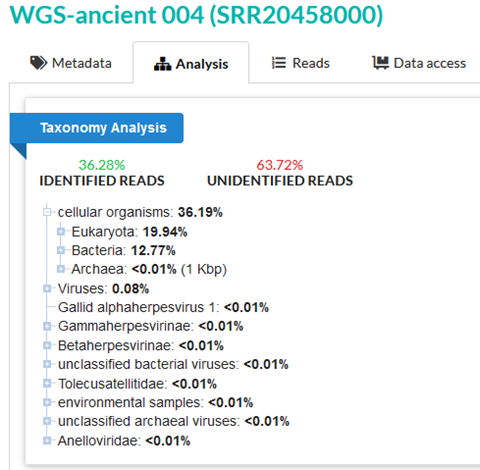
63.72% of reads in sample 4 are unidentified. This is most easily interpreted as a quality control issue of some kind – potentially caused by sample contamination, or very low-quality data.
The Abraxas report discusses the bioinformatics work that was done to match sample 4 reads to known genomes. Of note, 304,785,398 overlapped reads – a further processing step which the reads uploaded to SRA have not undergone – did not match to any of the tested genomes. However, after removing duplicate reads, this number was reduced by a factor of 10 to 30,823,217.
Continuing this analysis, they assembled the unique unknown reads for sample 4 into contigs. 65.69% of the unmapped reads were successfully assembled and re-matched to known organisms in the NCBI nt database. 97% of the assembled contigs were successfully matched to sequences in the nt database.
To summarize, the reads in sample 4 which could not be matched to tested species are on average highly duplicated reads. When duplicates were removed and the remaining unknown reads assembled into contigs, it resulted in the ability to match 64% of these remaining unknown reads to a database of known organism sequences.
The Abraxas report concludes with an acknowledgment that the NCBI nt database does not contain all sequences for all known organisms, and it is therefore certainly possible that the unidentified DNA reads are from already known (and therefore terrestrial) organisms which are not in the database.
The SRA taxonomy analysis figures still seem evocative, though – 64% unidentified? However, we can see that this is not unusual even for unambiguous ancient human DNA.
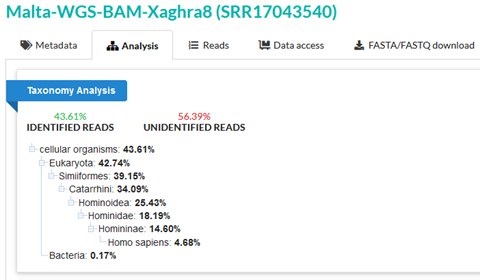
SRR17043540 is from a study into ancient Maltese genomes, and we can see that SRA taxonomy analysis gives 57% unidentified reads for this sample.
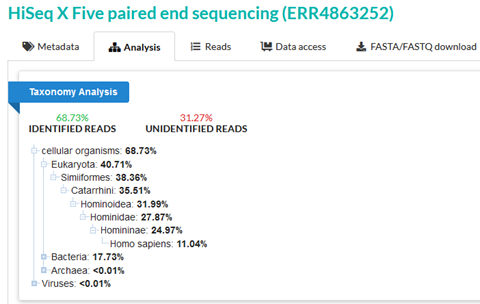
ERR4863252 is a sample from a single ancient human individual from the location corresponding to present-day France. Although the majority of reads in this sample are identified, 31.27% of reads are still unidentified by the SRA taxonomy analysis. And only 11.04% are confidently assigned as human.
Conclusion
So, after a review of the context surrounding the Nazca “alien mummies” and the genetic data presented as evidence of non-humanity – what conclusions can we draw? It seems clear that the genetic data is not conclusive evidence of non-human origins. Combined with the problems with the X-ray evidence espoused as proof of alien morphology – the Nazca mummies are not convincing. They may be assembled from ancient materials, but they are not ancient alien bodies.
Image Sources
[1] https://www.the-alien-project.com/en/mummies-of-nasca-alberto
[2] https://www.youtube.com/watch?v=7GVWup8KeQw
[3] https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&acc=SRR21031366&display=analysis
[4] https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&acc=SRR20755928&display=analysis
[5] https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&acc=SRR24975192&display=analysis
[6] https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&acc=SRR20458000&display=analysis
[7] https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&acc=SRR17043540&display=analysis
[8] https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&acc=ERR4863252&display=analysis