Is Biology Still a Wet Lab Science?
One of my friends recently published a research paper in one of the world’s most influential science journals, Science. [1] The paper is a significant milestone in our understanding of the neural crest, a strange group of cells in the vertebrate embryo with an unusual ability to mature into very different cell types pretty much anywhere in the body (cartilage, muscle, brain, skin, kidney, you name it). Evidence presented in the paper tackles a big unanswered question: how can these cells make precise choices on how to develop into, say, a neuron, in the absence of any significant external guidance? How does a precise and orderly sequence of differentiation choices emerge from the chaotic and random biochemical soup inside a neural crest cell?
My friend was a co-first author on the paper, which means that he picked up a bulk of the hard work that was involved in this breakthrough project. You probably imagine him to be a smart, hard-working guy in a lab coat with a pipette in his hand, moving mouse embryos from one Petri dish to another.
The only truthful thing in this picture is the smart & hard working bit! Ruslan, my friend, is not a biologist. Not even close. He’s only held a pipette once in his life! So how was he able to make a decisive contribution to a breakthrough biological discovery?
The short answer is that biology is no longer the same kind of science that it was for our parents and grandparents, and as such, it calls for a new type of scientist, like Ruslan. In order to give you the long answer, I need to put modern day biology in some historical context.
Low-Throughput Biology
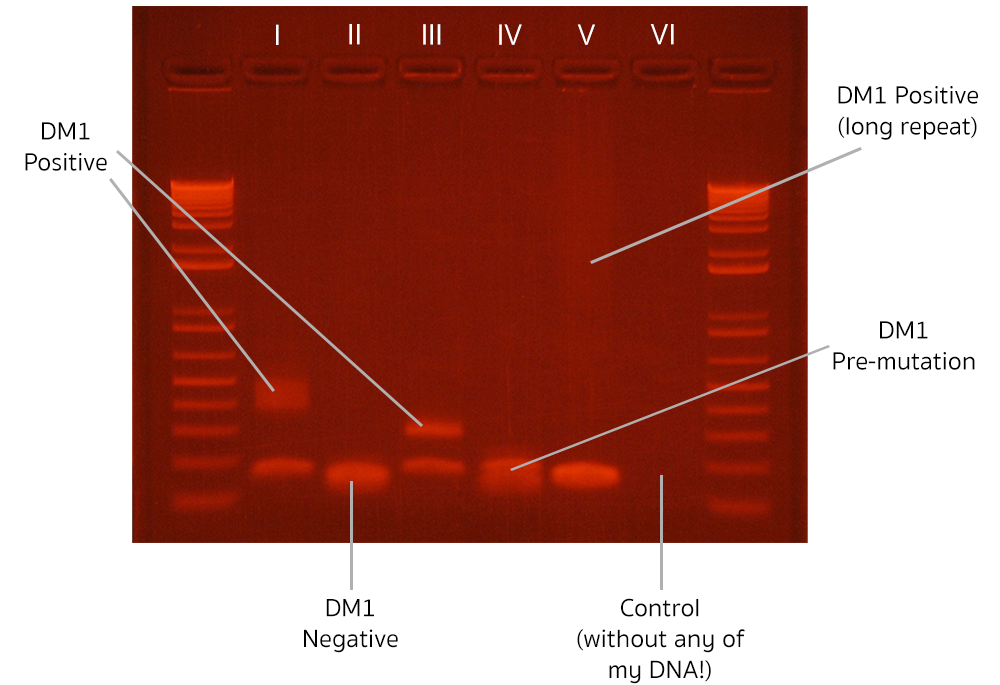
Biology used to be a low-throughput science, which means that a traditional biological experiment produces only a few data points. A good example of a low-throughput experiment is Polymerase Chain Reaction (PCR), a powerful technique of DNA amplification invented in the ‘80s. Amplified DNA is detected via gel electrophoresis. The whole experiment takes a couple of hours, and the outcome looks like this:
The measurements are fluorescent smudges (“bands”). The higher the position of the band is, the longer the piece of detected DNA. The more diffuse the band, the more unstable the length of the DNA is across different cells in the organism (this phenomenon is called “somatic mosaicism”; the faint, diffuse “long-repeat” measurement in column V indicates a high degree of somatic mosaicism). The first and the last columns contain a collection of DNA molecules with predetermined length, which act as reference points for the rest of the measurement (the “ladder”). As you can see, the whole experiment contains only a few measurements, which is the reason we classify it as “low-throughput”.
Despite low information density, low-throughput biology can be really powerful! The experiment presented above allowed me to diagnose three patients with a rare genetic disorder, Myotonic Dystrophy type 1 (DM1 positive: columns I, III and V; the expanded band in column V in faint and diffuse, so might be hard to notice), and diagnosed one patient with a pre-mutation version of the disease (DM1 pre-mutation: column IV). All while being very careful not to contaminate the experiment with my own DNA (negative control: column VI)!
Unfortunately, low-throughput biology is also really, really slow. I have a dual background in computer science/software engineering and biology, and I enjoy comparing these two very different fields. A skilled programmer can download an open-source web framework (e.g. node js), and create a Facebook clone in less than a day of work, including a database, login page, timeline, and ability to share photos. This means that iterating product ideas can be incredibly quick. Startup accelerator programs help software engineers to push this idea of quick iteration to the limits. Using short iteration cycles to quickly get customer feedback, hugely successful companies like Airbnb or Dropbox were created as summer projects.
Contrast this with biology. If things go smoothly, it takes 3 months to get permission from the ethics committee to do an experiment with human tissue. And even then, any progress faces substantial logistical challenges. Expensive reagents need to be shipped. Specialized equipment is often shared with other researchers in the lab and needs to be booked in advance. A tiny mistake in the beginning of the experiment may not be discovered until the final result is obtained, sometimes days or weeks later. Debugging information is often limited to “it didn’t work”. A single unit of measurable scientific progress (a scientific paper) takes ~100 low-throughput experiments. Each subsequent experiment incorporates learning from the previous experiment. The feedback cycle is measured in weeks to months. As a result it can take 2-5 years for a skilled researcher to write a scientific paper. A unit of externally measurable scientific progress, for example an effective drug, takes 50-100 scientific papers in the academic research phase, and up to 10 years of development in the biotech industry to bring the drug to market.
From the perspective of the ultimate “customer” of a research program (i.e. the patient) low-throughput biology delivers progress in the timescale of decades. Understanding and treating even a single-gene disease, like Myotonic Dystrophy is a task which can take an army of PhDs several generations. Understanding multi-gene diseases, like obesity or hypertension, is completely out of reach using low-throughput science.
High-Throughput Biology
High-throughput experiments are very different. A single high-throughput experiment allows a scientist to measure in the scale of millions or even billions of parameters at a time. Contrast that with just 10 measurements in the PCR experiment in the previous section. Economies of scale kick in, and the cost per measurement works out to be a tiny fraction of the cost per measurement of the corresponding low-throughput experiment. Why does it matter?
Let’s go back to the example I gave in the previous section. Using PCR we managed to diagnose several DM1 patients. The experiment cost my professor’s lab a few hundred USD and several hours of work of an experienced scientist. But what about Cystic Fibrosis (CF)? Or any other of close to 7000 rare diseases, which overall affect about 10% of the population? [2] If we decided to use low-throughput experiments to check all of them, we would quickly run out of budget and the scientist’s time. But it turns out that there’s a modern high-throughput biology experiment, which can detect more or less every single genetic disease like CF or DM1. It’s called Whole Genome Sequencing (WGS). In this type of experiment, the entire genetic material of a patient is measured. If you’re curious and like to self-experiment, you can even buy a direct to consumer WGS test online! [3]
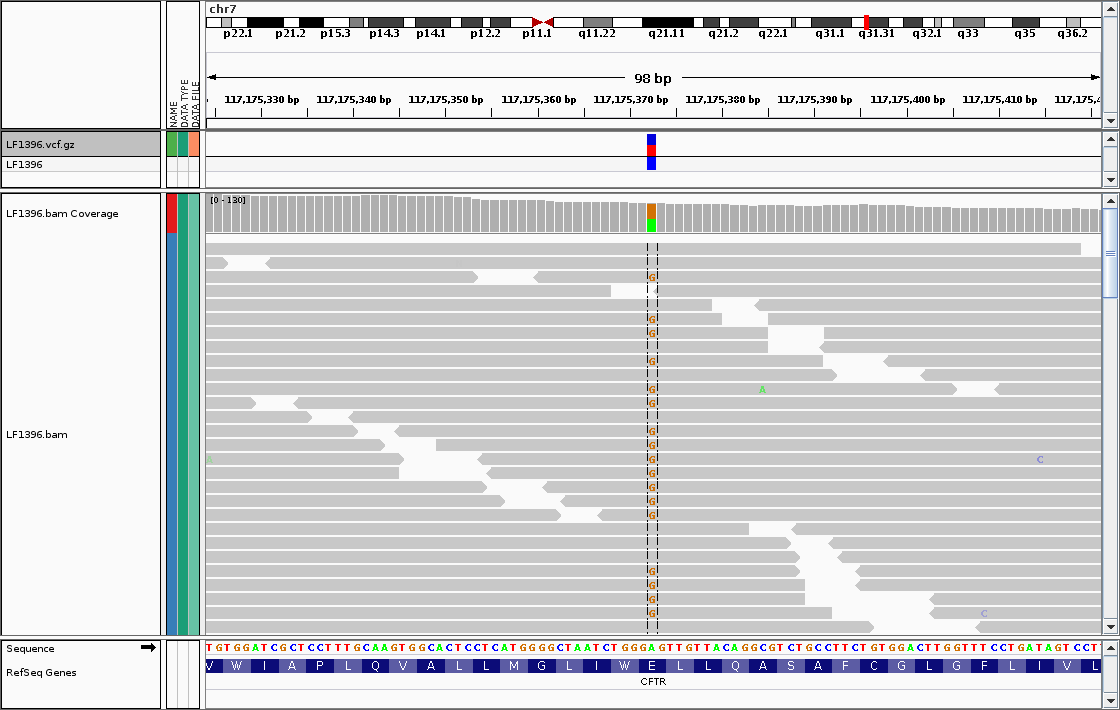
Unfortunately, it’s impossible for me to show here a picture of the entire WGS experiment. The amount of data is simply too large. In fact, it’s about 100 GB of data per sample — just like a 4k movie on Netflix! What is possible though is selecting a couple of measurements, and showing them on the screen. For example, the following picture shows how one can diagnose an uncommon variant in the Cystic Fibrosis gene.
Picture taken from prof. Jung Choi’s blog post.
Each grey, horizontal block is called a “read”. It is a basic unit of measurement in WGS. Each read is composed of nucleotides, basic building blocks of DNA. Nucleotides from the read, which are consistent with the “standard” human genome are grayed out. The anomalous nucleotides are highlighted in color. In this case, there is an anomalous nucleotide “G” in position 117,175,372 on chromosome 7. This variant indicates a mild CF phenotype, which presents with increased risk of bronchiectasis (see the original blog post for an extended discussion). [4]
Just to reiterate: this isn’t about just one genetic disease, like DM1 or CF. We can check the status of every single disease, that is, we can perform a comprehensive genetic screening test, quickly, cheaply and accurately.
High-throughput biology is now becoming ubiquitous. The virus which causes COVID-19 was first identified with a high-throughput method, viral genome sequencing. [5] A new type of sequencing (“nanopore sequencing”) can be used to perform an unbiased diagnosis of any pathogen (virus, bacteria or fungus) within hours from taking a blood sample [6,7, 8]. This technology will become a real game changer in diseases where prompt diagnosis is required to administer a correct, life-saving medication. Going back to basic research, just one single-cell sequencing experiment is now enough to derive a detailed understanding of the process of blood production (hematopoiesis), a scientific discovery process that took decades beforehand [9].
It needs to be noted that high-throughput biology still hasn’t replaced low-throughput biology, and perhaps never will. While it’s true that the per-measurement cost of a high-throughput experiment is low, the total cost of the experiment is usually at least an order of magnitude higher than that of a corresponding low-throughput version. This matters, if the scientist cares about 10 measurements only and not the remaining 999,990. And there are still entire classes of experiments, which are only available in the low-throughput version (e.g. most in vivo biology, like microsurgery, xenograft tumor models, etc.). Low-throughput experiments can also be tweaked and customized for non-standard applications, and are easier to “debug”. That’s why the majority of biology papers are still published with low-throughput experiments as the major source of data.
Bioinformatics
High-throughput biology presents new challenges that weren’t a big part of biology before. Specifically, it requires extracting insights from large, messy, noisy datasets. The basic skill set is no longer that of careful manipulation of small volumes of liquids, breeding animals, and orchestrating a series of low-throughput experiments. The basic skillset is programming, data cleaning, data manipulation, and statistical modeling. The actual wet lab science of performing a high-throughput experiment quickly becomes standardized and is often outsourced through core facilities or CROs!
What is left is making sense of a large, detailed dataset. It still involves considerable knowledge of biology and requires biological intuition necessary to reason about a very complex system (a cancerous tumor, visual cortex, immune system or RNA splicing disease mechanism) with poor observability. Even with a high-throughput technique we can only measure a tiny fraction of the overall system, omitting many important details. High-throughput biology requires a new type of scientist, like Ruslan. It requires a bioinformatician.
What Are the Stakes?
The stakes are high. Precision medicine in cancer is a natural application of this skillset. Tempus, precision medicine company, is reportedly sequencing every third cancer in the US. The information that a patient can learn from their sequencing report can already be life saving [10]. And it will get only better as Tempus’ dataset grows larger and their algorithms get better at highlighting the best treatment options.
It is only a matter of time before Polygenic Risk Scores, computed from massive genetic datasets like the UK Biobank, will become a standard tool in clinical practice, helping to more quickly identify patients at risk of diseases like coronary heart disease and breast cancer [11].
Sequencing the COVID-19 genome helped to identify outbreak clusters by tracing infected individuals to specific places, like a church or a hospital. [12] A system built around this technology could help us more successfully manage future pandemics.
So is biology still a wet lab science? At its core it still is. Every biological question is answered by looking at data generated in the lab. But with explosive growth in the size and complexity of that data, “looking at the data” is often the creative, difficult step, which makes or breaks a breakthrough science project.
About the author
Adam Kurkiewicz is a computer programmer, bioinformatician, angel investor, entrepreneur, and venture scout at Backed.vc. He is also the CEO of a biologist-friendly data analytics company Biomage. There, he co-developed Cellscope, a single cell RNA-seq data analysis tool, which is currently used at top universities world-wide, including Harvard Medical School, Stanford, and University of Oxford. You can reach him at adam@biomage.net.
References
[1] https://science.sciencemag.org/content/364/6444/eaas9536
[2] https://www.genome.gov/FAQ/Rare-Diseases#:~:text=A%20rare%20disease%20is%20generally,million%20to%2030%20million%20Americans
[3] https://www.mobihealthnews.com/news/nebula-genomics-launches-299-direct-consumer-whole-genome-sequencing
[4] https://jchoigt.wordpress.com/2012/07/18/working-with-23andme-exome-data-my-cf-allele-and-the-need-for-verification/
[5] https://www.nejm.org/doi/full/10.1056/nejmoa2001017
[6] https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-015-0220-9
[7] https://doi.org/10.1038/s41587-019-0156-5
[8] https://pubmed.ncbi.nlm.nih.gov/26823918/
[9] https://science.sciencemag.org/content/367/6479/eaaw3381?rss=1
[10] https://youtu.be/QH1OORy4jsI
[11] https://www.nature.com/articles/s41588-018-0183-z
[12] https://www.nature.com/articles/s41598-021-85623-6